Overview
PetriNet is an Add-on to handle discrete and continuous place-transition nets of varying complexity. The editing of Petri nets and the transformation of biological networks into Petri nets is possible in an interactive way. Simulation and analysis are supported by different visualization and interaction techniques to capture the properties and behaviour of Petri nets quickly (e. g. interactive animation of the switching behavior, interactive exploration of invariants and the reachability graph).Please cite the following paper (1) or master thesis (2) if you use PetriNet:
Hartmann, A., H. Rohn, K. Pucknat & F. Schreiber:
Petri Nets in VANTED: Simulation of Barley Seed Metabolism.
Proceedings of the 3rd International Workshop on Biological Processes & Petri Nets (BioPPN), CEUR Workshop Proceedings Volume 852 (2012) 20-28.
Kevin Pucknat (2012)
Erweiterung der Software VANTED zum Editieren und Simulieren biologischer Petri-Netze
(master thesis, Extension of the software VANTED for editing and simulation of biological Petri-Nets)
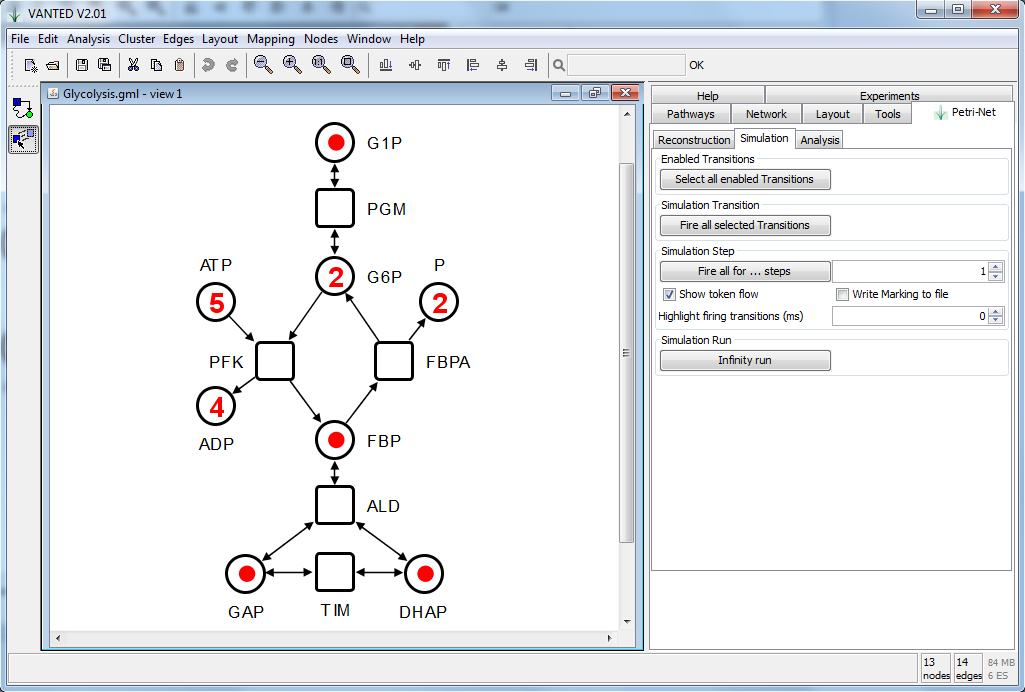
Figure 1.: Screenshot of VANTED and the Petri-Net
add-on visualizing the glycolysis pathway as a Petri net.
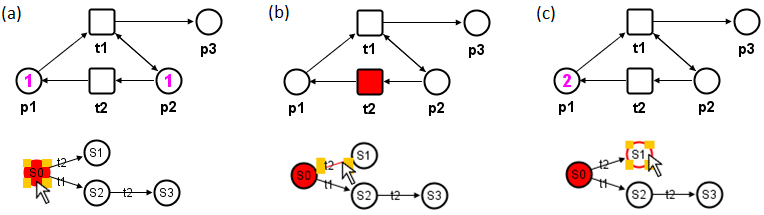
Figure 2.: Interactive visualization of the Petri net reachability analysis. (a) Initial marking
is represented by the initial state S0 (marked) of the reachability graph, which is
visualized in the Petri net. (b) Mouse-over an edge of the reachability graph results
in a visualization of the firing transition (t2), which would lead to the next reachable
state S1 (c).